However, a new paper in Nucleic Acids Research reminds us that intermediary forms are not always necessary in evolution. Resch and Striegl et al. [36(13):4390-4401] report a sudden switch in function in the evolution of a regulatory protein, which changes between two mutually exclusive modes of action with just one amino acid change. No conceptually impossible intermediate necessary!
Before I get into the paper, let me back up and go over some background you’ll need to understand it. The paper focuses on a bacterial repressor protein. In bacteria, repressor proteins stop transcription of a gene by binding to a certain spot in DNA, called the operator. Let me explain. Normally, for DNA’s information to be used, it first has to be transcribed into RNA by RNA polymerase. RNA polymerase binds to the DNA and travels along it, letting out a growing string of newly-transcribed RNA as it passes along. Before beginning to transcribe the actual gene in the DNA, however, the RNA polymerase must pass over the operator. If nothing is bound to the operator, the RNA polymerase can just continue on and complete the transcription. If, however, a repressor protein is bound to the operator, it physically blocks the RNA polymerase from passing, thus preventing the gene from being transcribed and “read.” (If you’re not a biology person and can’t picture this process blindfolded with your hands tied behind your back, check out this video.
There’s an added level of complexity when you consider what is regulating (controlling) the repressor. There are two completely opposite ways of doing this. One is through an inducer, which is a molecule that binds to a repressor, preventing it from binding to the operator. The second is through a co-repressor, which binds to the repressor, helping it bind to the operator. So, inducer → gene expression, co-repressor → no gene expression. Two totally different options. To illustrate, see the pictures and captions below:
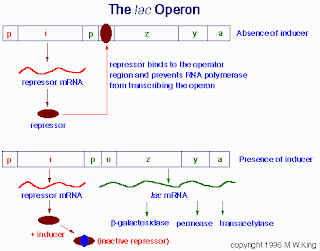 This is a set of genes called the Lac Operon, which is regulated by a repressor and an inducer. Notice how the repressor (maroon oval) is always bound to the operator to prevent transcription, unless the inducer (blue oval) attaches to the repressor causing it to fall off and allowing transcription to occur.
This is a set of genes called the Lac Operon, which is regulated by a repressor and an inducer. Notice how the repressor (maroon oval) is always bound to the operator to prevent transcription, unless the inducer (blue oval) attaches to the repressor causing it to fall off and allowing transcription to occur.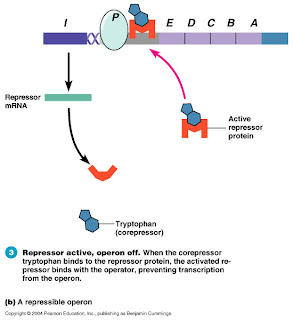 This one is a picture of the Trp Operon, regulated by a repressor and a co-repressor. Normally, the repressor (U-shaped orange thingie) is not bound to the operator, and gene transcription occurs. However, when the co-repressor (blue hexagon+pentagon) is present, it binds to the repressor, allowing it to bind to the DNA, stopping transcription.
This one is a picture of the Trp Operon, regulated by a repressor and a co-repressor. Normally, the repressor (U-shaped orange thingie) is not bound to the operator, and gene transcription occurs. However, when the co-repressor (blue hexagon+pentagon) is present, it binds to the repressor, allowing it to bind to the DNA, stopping transcription.The take-home message from this is that repressor-mediated gene regulation can occur in two opposite ways: two things binding together to stop transcription, or two things binding together to allow transcription.
In the paper by Resch and Striegl et al., they describe how one regulator switched between the two opposites listed above in just one small evolutionary step. The regulator in question is called TetR, pictured below, bound to its operator on a strand of DNA:

TetR is involved in tetracycline resistance in bacteria. (Bacteria have a habit of quickly evolving resistance to whatever antibiotics we throw at them, and they’ve done particularly well with tetracycline—there is not just one but at least three different mechanisms for tetracycline resistance in bacteria. For an interesting review, check out this paper by Speer et al. from 1992. TetR works like the regulator in the Lac operon in the example above. Its normal position is to be bound to the DNA, preventing transcription of its tetracycline-resistance genes. Antibiotic resistance is often costly for bacteria, and they generally only activate their resistance when it’s needed. In this case, the resistance is needed when tetracycline is present in the environment. The tetracycline acts as the inducer, binding to TetR, which causes it to release from the DNA, allowing transcription of the resistance genes. When tetracycline is removed from the environment, TetR is able to bind to the DNA again.
The weird thing is, there’s several mutant versions of Tet-R (called revTet-R) that have exactly the opposite behavior. revTet-R cannot bind to DNA unless tetracycline is present, much like in the example of the Trp operon, above. In the mutant, tetracycline suddenly acts as a co-repressor, not an inducer. (This results in tetracycline in the environment turning off the tetracycline-resistance genes, and in the genes being on once tetracycline goes away.) Even weirder, one of these mutants is only one amino acid different than the sequence of the original Tet-R—the 17th leucine in the chain is replaced by a glycine. How could a change of just one measly amino acid in this large, complex protein change induce a completely opposite behavior from the one it originally possessed?
The authors and their team used a variety of biochemical procedures to answer this question. They raised the revTet-R mutant bacteria, broke open their cells, purified the contents to get out the revTet-R protein, broke it up, then sequenced the bits. Then they determined its three-dimensional structure with and without attachment to tetracycline.
Here’s a picture of the two molecules taken from the article, going through their processes of binding tetracycline and DNA, side-by side. The line drawings represent the overall structure of Tet-R and its variant. The left-most, vertical arrangement (A-C) is revTet-R, and the horizontal arrangement (D-F) is of Tet-R. The “effector” is tetracycline, and it binds to that little oval in the center of the egg-shaped part of the protein.
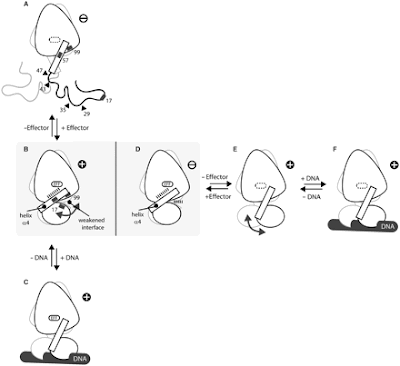
Let’s start with the standard Tet-R, images D-F. Notice that the helix α4 (the rectangle) functions like a pendulum, swinging up and tightening in the two ball shapes tight in close to the main egg-shape when tetracycline (the effector) is present. This prevents DNA from being able to fit into the binding site to attach. But in the absence of tetracycline, the pendulum swings down, opening up the binding site to DNA. However, in revTet-R, in the absence of tetracycline (A) the amino acid chains that are normally rolled up into two balls no longer want to bind up together. Instead, they are loose and free, and don’t make any of the three-dimensional structure necessary to bind DNA. The slight change in amino acid sequence in the mutant is enough to screw up the whole DNA binding apparatus. However, when tetracycline is bound in (B), all of a sudden everything tightens up, and the result looks a lot like (D): Tet-R with tetracycline. But, because of that amino acid substitution, the normally tightly-bound DNA binding site is not as tight as it usually is. This weakness allows DNA entry into the binding site even when tetracycline is attached, resulting in revTet-R to have an opposite action to Tet-R.
So from just one single amino acid change, this protein called Tet-R completely reverses its function, but through a completely different mechanism and set of conformational changes than it goes through normally. This just wows me with its level of specificity—with all the thousands of amino acids in this protein, just one change can screw everything up in such a drastic way. Not just to break it—which is easy—but actually to reverse it through a whole new mode of action. I’d imagine that creationist folks would say this is evidence of the delicacy of life, how everything is too fragile and precise to have arisen on its own without being designed. But I would argue that this incredible ability to change gears is evidence for the power of random change—how even if an organism’s environment suddenly does a 180, random mutation is sufficient to allow it to go with the flow and allowing the species to adapt in a radical manner to whatever its ecosystem can throw at it. Obviously in this case, a revTet-R mutant doesn’t seem very well-adapted to a bacterium’s normal environment. Turn off your tetracycline-resistant genes in the presence of tetracycline? (It was generated in a laboratory through random mutagenesis, not evolved naturally in the wild.) But it’s a great living analogy for the power of mutation to do crazy, seemingly-impossible things. Obviously evolution often occurs through slow steps and many intermediates, especially for larger, more complex organisms. But this paper shows that it doesn’t always have to happen that way.

No comments:
Post a Comment